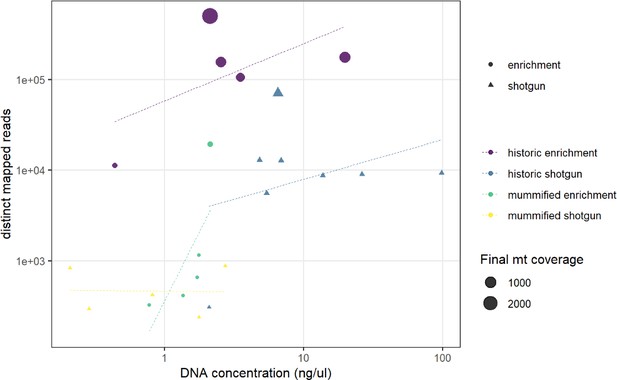
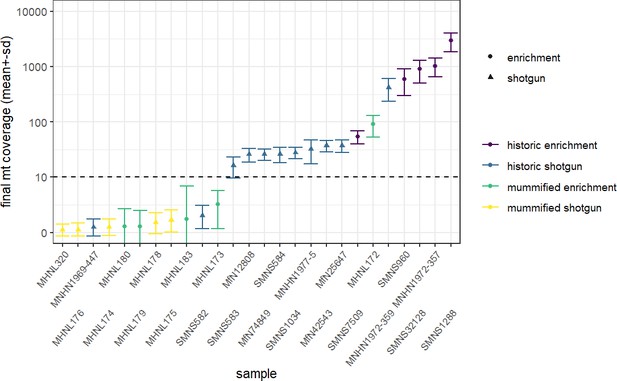
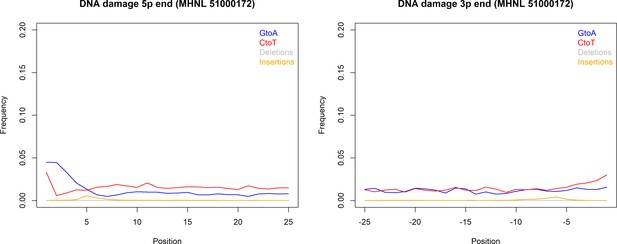
We succeeded in sequencing the mitogenomes of 14 historic baboons from northeastern Africa and a mummified baboon recovered from Gabbanat el-Qurud, presenting the first genetic data of a mummified baboon from ancient Egypt to date. DNA of the mummified baboon shows post-mortem damage, which is, however, relatively low compared to what can be expected for samples of similar age (Dabney et al., 2013b, Kistler et al., 2017). Low frequencies of post-mortem damage were observed for aDNA from mummified specimens and have been attributed to the water deprivation during the mummification process, which may prevent hydrolytic deamination (Rossi et al., 2021). Post-mortem damage observed here is within the range previously reported for aDNA derived from mummified Egyptians (Schuenemann et al., 2017) and sheep recovered from an Iranian saltmine (Rossi et al., 2021), which supports the authentic origin of our ancient sequence data and tends to rule out the possibility of contamination with modern DNA. The very low frequency of mismatches in the mapped reads from the mummified sample and its unique sequence are further evidence against the concern of contamination from other baboon samples.
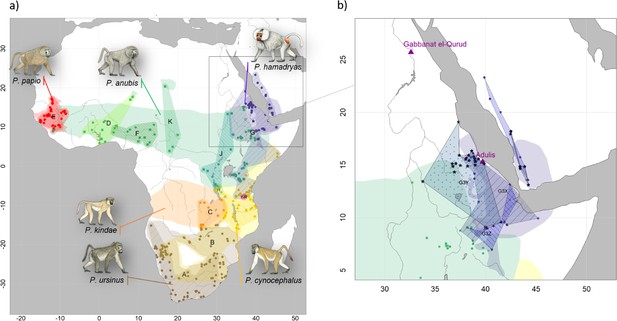
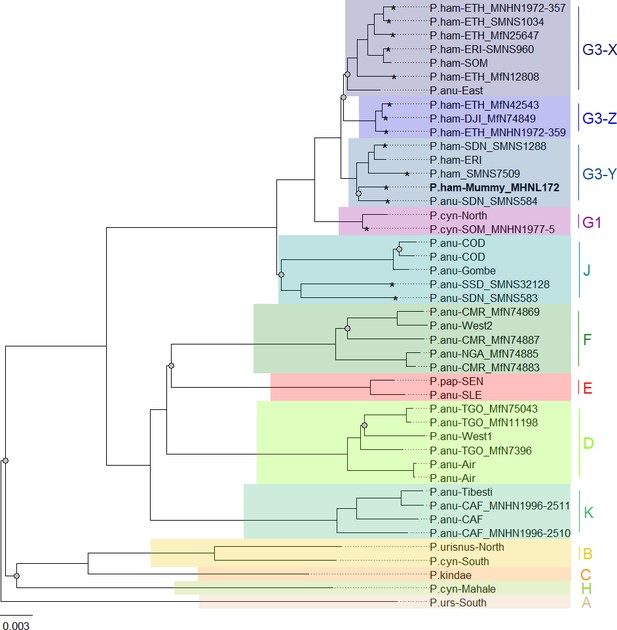
Our phylogenetic analysis of the newly generated mitogenomes in combination with published mitochondrial sequence data produced tree topologies in agreement with those of prior studies, with three well-supported clades across the northeastern distribution of Papio (Roos et al., 2021). As previously described, introgressive hybridization has led to discordances between species assignment and mitochondrial clades (Rogers et al., 2019; Sørensen et al., 2023; Zinner et al., 2009; Zinner et al., 2011). Our findings are notable for including specimens from previously unsampled or underrepresented regions, filling gaps in our knowledge of the distribution of mitochondrial clades. For instance, we report mitochondrial sequence data of baboons from regions previously unstudied, namely South Sudan and Sudan. We show that samples from South and southern Sudan, east of the White Nile, nest within the central olive baboon clade J, whereas samples from the coastal region of Sudan and east of the Blue Nile nest within the hamadryas clade G3. These findings expand the northern distributions of both clade J and clade G3 significantly, while also highlighting a strong geographic affinity between clade J and the Albertine Rift and (White) Nile Valley. Taxonomically, this clade corresponds with two subspecies recognized by Hill, 1970: P. a. heuglini and P. a. tesselatum.
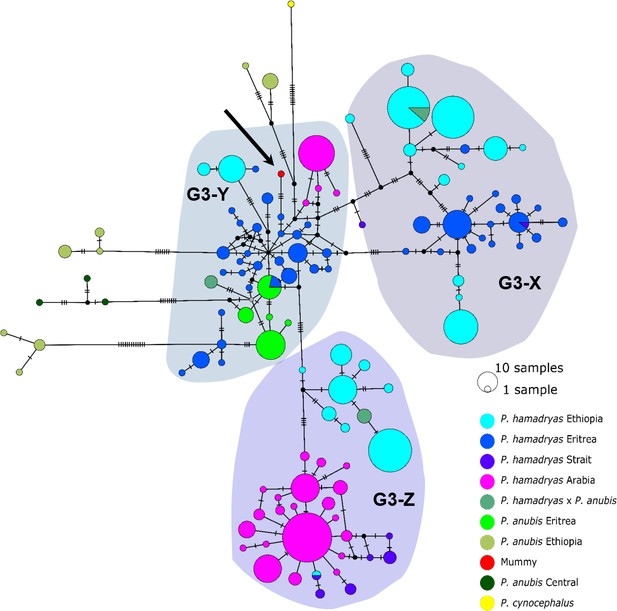
A mummified hamadryas baboon from Gabbanat el-Qurud (MHNL 51000172) yielded sufficient aDNA to produce a complete mitogenome, which fell unequivocally in subclade G3-Y (cf. Kopp et al., 2014b). Haplotype networks allowed us to further refine subclade G3-Y, which consists of P. hamadryas and P. anubis samples from Eritrea and P. anubis samples from neighbouring regions in Sudan. G3-Y also includes samples from the southern-most distribution of P. hamadryas on the Arabian Peninsula. Geographic stability of phylogenetic clades over millennia has been shown for other baboon populations (Mathieson et al., 2020), leading us to infer that MHNL 51000172 (or its maternal ancestor) originated in the region where clade G3-Y exists today. We cannot completely rule out an Arabian origin for MHNL 51000172, as our data does not cover the entire historic and present haplotype diversity there, but the tight clustering of the currently available Arabian sequences and distances in the HVRI network make an Arabian origin of MHNL 51000172 unlikely. Similarly, the close relationship with a sample of P. anubis from Sudan east of the Blue Nile (SMNS-Z-MAM-000584) could indicate trafficking of baboons along the Nile, as suggested for specimens of P. anubis recovered from Ptolemaic catacombs (Brandon-Jones and Goudsmit, 2022; Peters, 2020) and the Predynastic site of Hierakonpolis (Van Neer et al., 2004). However, MHNL 51000172 was identified phenotypically as P. hamadryas (Lortet and Gaillard, 1907), and the distribution of hamadryas baboons is restricted to more eastern regions (Figure 2). If the distributions of baboons in northeastern Africa have remained roughly stable within the last 2500 y (as supported by ecological niche modelling; Chala et al., 2019), the region in Sudan east of the Blue Nile and west of the Atbarah River could not have served as a source region for hamadryas baboons. Thus, it stands to reason that MHNL 51000172 (or its maternal ancestor) was captured in present-day Eritrea (or close neighbouring regions) and trafficked to Egypt. The value of this finding is twofold. First, it connects the mummified baboon to populations that live today in Eritrea and eastern Sudan, between 13° and 20° latitude. Second, it corroborates the reports of Greco-Roman historians, who described Eritrea, and specifically Adulis, as the sole source of P. hamadryas for Ptolemaic Egyptians.
Yet, this baboon predates the reign of Ptolemy I by centuries, presuming it is contemporaneous with another baboon (MHNL 90001206) in the same assemblage, ca. 800–540 BCE (Richardin et al., 2017). Thus, our findings raise the possibility that Adulis already existed as a trading centre or entrepôt during the 25th and 26th dynasties of Egypt. Although speculative, and expressed with due caution, our reasoning would extend the antiquity of Egyptian–Adulite trade by as much as five centuries.
Arguing for pre-Ptolemaic contact between Egypt and Adulis is fraught in the absence of corroborating material evidence—but even so, the archaeological record is not entirely silent on the prospect. Manzo, 2010 and others (Zazzaro et al., 2014) reassessed the ceramic tradition at Adulis and developed a chronology that stretches to the early second millennium BCE, the deepest levels of which contained a fragment of blue glass with yellow inlays similar to Egyptian glass from the New Kingdom (Fattovich, 2018). In Egypt, contact with the Eritrean lowlands is attested by trade goods dating to ca. 1800–1650 BCE or earlier, including potsherds, obsidian, and fragments of carbonized ebony (Fattovich, 2018; Lucarini et al., 2020). Discovered at Mersa Gawasis, a Middle Kingdom harbour, these artefacts appear to align the prehistory of Adulis with the fabled Land of Punt (Bard and Fattovich, 2018; Manzo, 2010; Manzo, 2012), an enigmatic toponym scattered across scant and disconnected records (Cooper, 2020).
Punt existed in a region south and east of Egypt, and was accessible by land or sea. For Egyptians, Punt was a source of 'marvels,' particularly incense, but also baboons, that drove bidirectional trade for 1300 y (ca. 2500–1170 BCE) (Tallet, 2013). Some scholars have described this enterprise as the beginning of economic globalization (Fattovich, 2012), whereas others view it as the earliest maritime leg of the spice route (Keay, 2006), a trade network that would shape geopolitical fortunes for millennia. The global historical importance of Punt is therefore considerable, but there is a problem—its location is uncertain, in part because the toponym fades from view. From the early first millennium BCE, there are no further records of Egyptians in Punt or of Puntites visiting Egypt. There are, however, two incomplete inscriptions that mention Punt in a narrative context, and both are attributed to the 26th (Saite) Dynasty (Betrò, 1996; Cavasin, 2019). One of these, the Defenneh stele, describes an expedition to Punt that was saved from dying thirst by unexpected rainfall on 'the mountains of Punt' (Meeks, 2003). The Defenneh stele is a testament to the efforts of Saitic pharaohs to revive maritime commerce on the Red Sea (Lloyd, 1977), while also raising the possibility of renewed trade with Punt. It is perhaps no coincidence that the Saite dynasty (664–525 BCE) exists squarely within the radiometric date range of hamadryas baboons from Gabbanat el-Qurud.
Punt, like Adulis, was a source of baboons for Egyptians, a history that raises the possibility of using baboons as a tool for testing geographic hypotheses. Recently, Dominy et al., 2020 used stable isotope mapping methods to determine the geoprovenance of mummified baboons from Thebes (modern-day Luxor) and dated to the (late) New Kingdom. Their results pointed to present-day Ethiopia, Eritrea, or Djibouti, as well as portions of Somalia, an area that corroborates most scholarly views on the location of Punt (Breyer, 2016; Kitchen, 2004), but see Meeks, 2002; Meeks, 2003; Tallet, 2013. Here, we used aDNA to show that at least one baboon from the 25th Dynasty or Late Period of Egyptian history—a span that coincides with the last known expeditions to Punt, but predates Greco-Roman accounts of Adulis as a source of baboons—can be traced to Eritrea. Thus, our findings appear to establish primatological continuity between Punt and Adulis. Such a conclusion must be viewed with caution, but it bolsters recurrent conjecture among some historical archaeologists: that Punt and Adulis were essentially the same trading centre from different eras of Egyptian antiquity (Doresse, 1959; Fattovich, 2018; Kitchen, 2004; Massa, 2021; Phillips, 1997; Sleeswyk, 1983).
At minimum, our results reinforce the view that ancient Egyptian mariners travelled great distances to acquire living baboons. A great strength of this conclusion is that it is based on distinct but complementary methods, but of course, the sample size is paltry and limited to P. hamadryas, one of two baboon species recovered from Gabbanat el-Qurud. Moving forward, it would be desirable to expand the sample size, examine specimens of P. anubis as well as nuclear genomic data for increased precision, and include different time intervals of baboon mummification.
Direct radiocarbon dating of MHNL 51000172 and other baboons from Gabbanat el-Qurud is an urgent priority, in part because doing so would put these specimens into conversation with those from the catacombs of Tuna el-Gebel. The oldest gallery at Tuna el-Gebel, Gallery D, is dated to the 26th Dynasty and contains a single species of baboon: P. anubis. Some scholars (Peters, 2020; von den Driesch et al., 2004) have argued that these olive baboons, as well as Chlorocebus aethiops (also found in Gallery D), were sourced from the Sudanese Nile Valley and adjacent areas, which predicts membership in clade G3-Y, although clade J is also plausible. Construction of Gallery C began during the first period of Persian rule in Egypt (524–404 BCE) and continued through the 30th and Ptolemaic dynasties. As every phase of Gallery C contains mummified specimens of both P. anubis and P. hamadryas, there is rich opportunity to explore diachronic changes in trade routes using phylogeographic methods. Uniform membership in clade G3-Y, for example, would affirm that Late Period Egyptians were sourcing P. hamadryas from Eritrea as early as the sixth century BCE. Testing this hypothesis may prove rewarding.